Sequences equivalent to their reverse complements (i.e., double-stranded DNA) have no equivalent in text analysis and non-biological string algorithms. Despite this striking difference, algorithms designed for computational biology (e.g., sketching algorithms) are designed and tested in the same way as classical string algorithms.
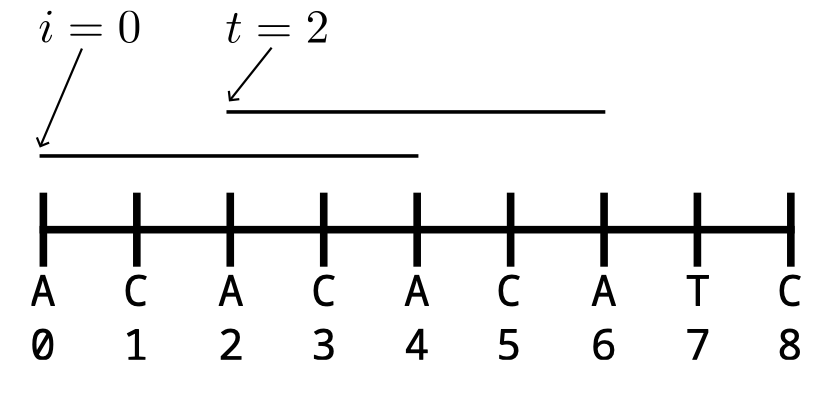
Then, as a post-processing step, these algorithms are adapted to work with genomic sequences by folding a k-mer and its reverse complement into a single sequence: the canonical representation (k-nonical space). The effect of using the canonical representation with sketching methods is understudied and not understood.
As a first step, we use context-free sketching methods to illustrate the potentially detrimental effects of using canonical k-mers with string algorithms not designed to accommodate for them. In particular, we show that large stretches of the genome (“sketching deserts”) are undersampled or entirely skipped by context-free sketching methods, effectively making these genomic regions invisible to subsequent algorithms using these sketches. We provide empirical data showing these effects and develop a theoretical framework explaining the appearance of sketching deserts. Finally, we propose two schemes to accomodate for these effects: (1) a new procedure that adapts existing sketching methods to k-nonical space and (2) an optimization procedure to directly design new sketching methods for k-nonical space.