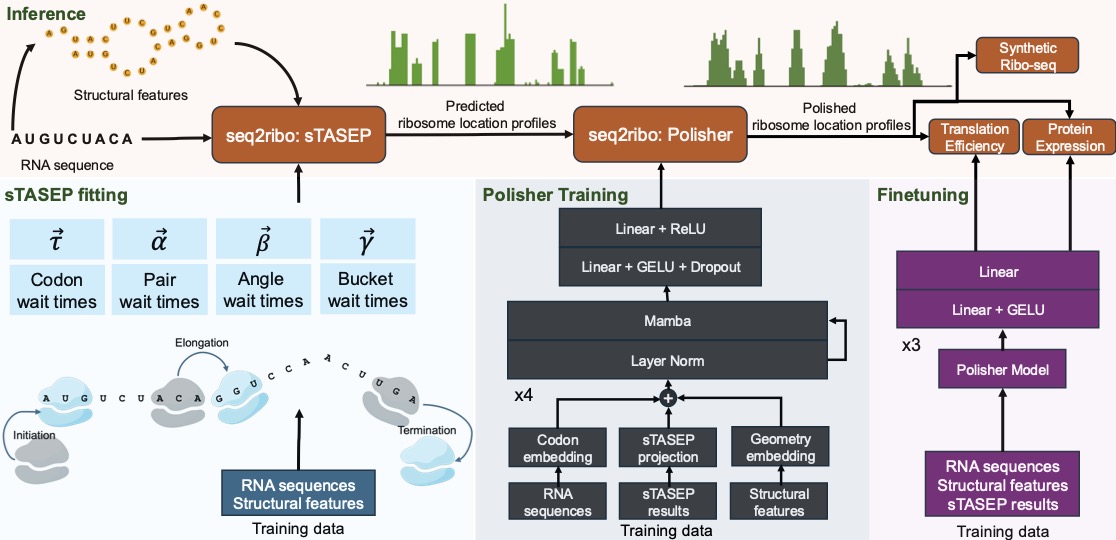
Motivation: Ribosome dynamics are vital in the process of protein expression. Current methods rely on ribosome profiling (Ribo-seq), RNA-seq profiles, and full genomic context. This restricts their use in de novo sequence design, like messenger RNA (mRNA) vaccines. Simulation-only approaches like the Totally Asymmetric Simple Exclusion Process (TASEP) oversimplify translation by focusing solely on codon elongation times.
Results: We present seq2ribo, a hybrid simulation and machine learning framework that predicts ribosome A-site locations using only an mRNA sequence as input. Our method first employs a novel structure-aware TASEP (sTASEP), which models translation using a comprehensive set of fitted parameters that include codon wait times and structural features, such as local angles, base-pairing, and discrete positional buckets. The ribosome locations generated by sTASEP are then processed by a polisher model, which learns to refine the simulated ribosome distributions. seq2ribo provides high-fidelity predictions of ribosome locations across diverse cell types (iPSC, HEK293, LCL, and RPE-1), significantly outperforming baselines. When benchmarked against sequence-only Translatomer, seq2ribo achieves reductions in transcript-level error up to 35.8%, while simultaneously attaining the highest Pearson and Spearman correlations in every cell line and reducing structural errors between 43.3% and 97.3%. By adding a task-specific head, seq2ribo achieves Spearman correlations up to 0.795 with experimental translation efficiency (TE) across several cell lines, and 0.689 with measured protein expression. By operating from sequence alone, seq2ribo provides a new tool for synthetic biology, enabling the rational design and optimization of mRNA sequences without the need for expression-level data or genomic context. Availability: seq2ribo is available at https://github.com/Kingsford-Group/seq2ribo. Contact: gkaynar{at}cs.cmu.edu, carlk{at}cs.cmu.edu. Supplementary information: Supplementary data are available.